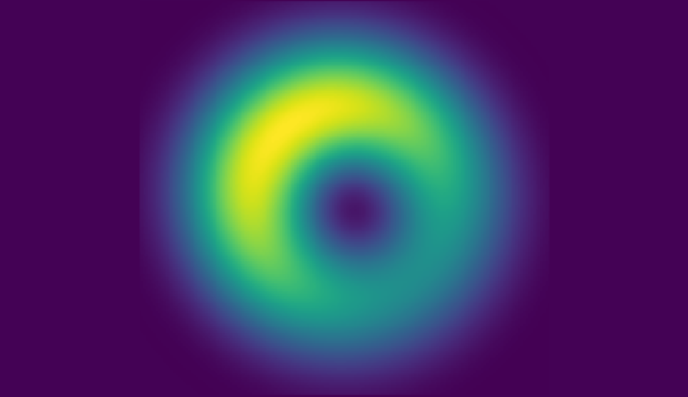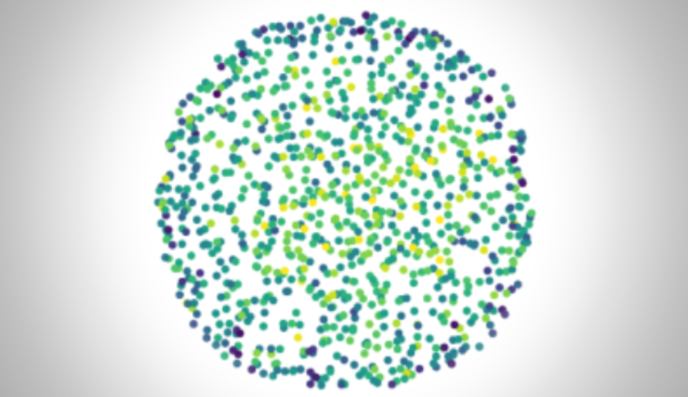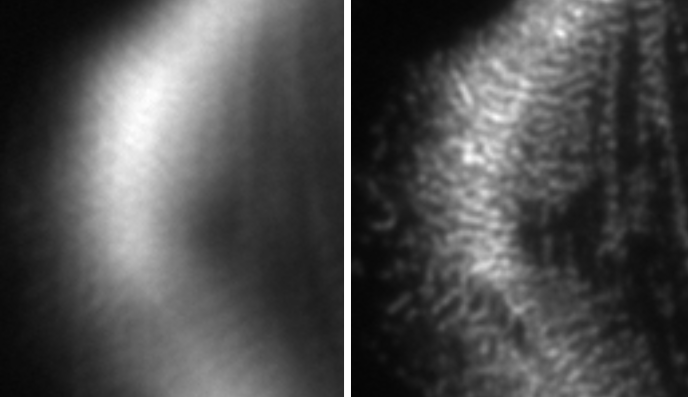
Microscopy images are often distorted due to optical imperfections or inhomogeneities within sample. In collaboration with the
Shroff lab, I work on enhancing wavefront sensing and image restoration using both classical and machine learning methods, with the aim of achieving optimal correction in real-time.
Analytical method: [Paper]
[Code]
DeepPD:
[Preprint]